Comparative Functional Genomics Lab
Project title
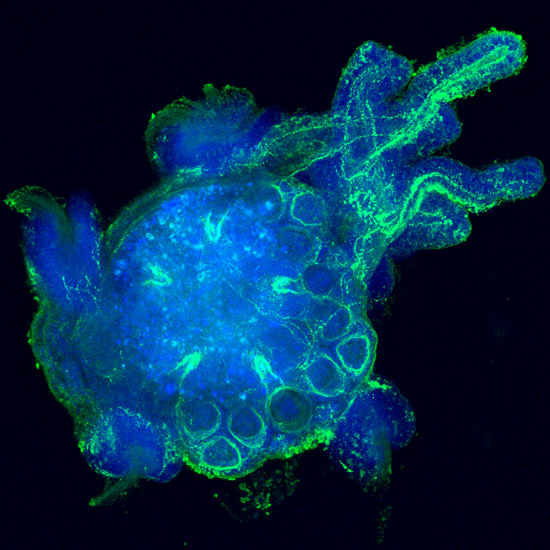
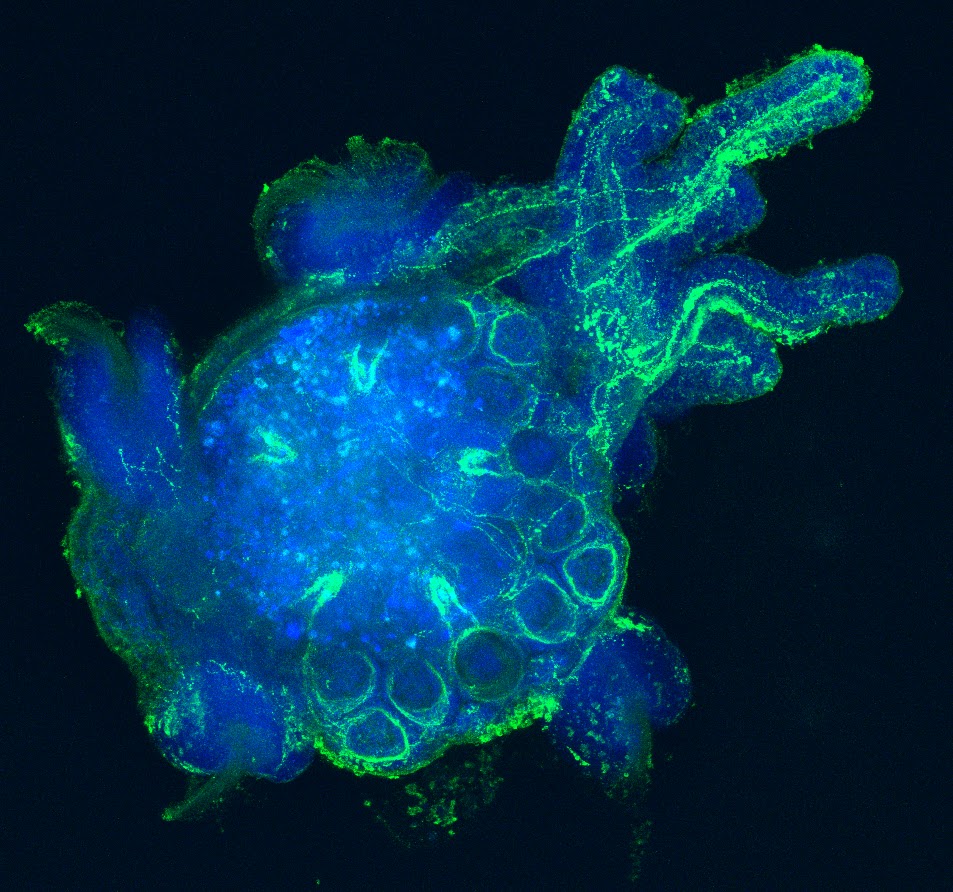
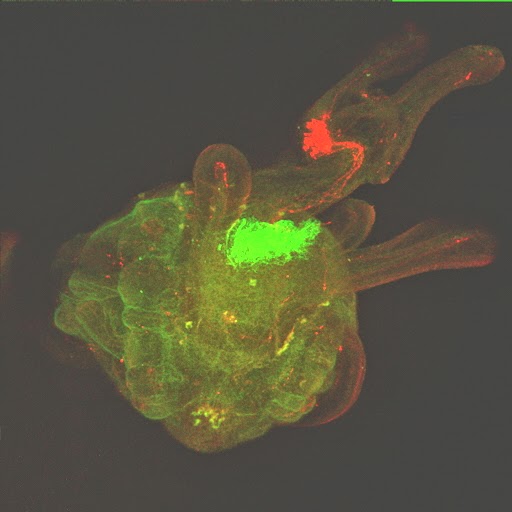
Over the past years, my laboratory has generated novel comparative and genomic data related to the large scale characterization of transcriptomes from a variety of organisms representing major animal lineages such as Lophotrochozoa (i.e. Aplysia californica and related molluscs (Heyland et al., 2011b; Moroz et al., 2006), basal Deuterostomes (Xenoturbella boci) (Bourlat et al., 2006)Placozoa (Trichoplax adherens) (Heyland et al., in press)and Ecdysozoa (Flatt et al., 2006; Neira Oviedo et al., 2009). Specifically, we used standard and innovative massive parallel sequencing systems (e.g. 454 Life Sci pyrosequencing, Illumina) to obtain datasets contributing to functional genomics approaches in a diversity of species. Work with transcriptomes has not only helped to clarify the position of these organisms within metazoan phyla (for Xenoturbella) (Bourlat et al., 2006)but also revealed many new interesting neuropeptides and transcription factors that appear to have been evolutionarily lost in the ecdysozoan or vertebrate model systems (Bourlat et al., 2006; Heyland et al., 2011b; Moroz et al., 2006). My lab’s contributions to these projects were i) generating representative cDNA libraries which were used for high throughput sequencing technologies, ii) application of a diverse array of phylogenetic methods, iii) database work and manual annotation of signal transduction pathways, iv) microarray experiments and analysis v) training HQP at various levels in functional genomics and bioinformatics (Heyland et al., 2012; Heyland et al., 2011b)and vi) immunohistochemical and in situ hybridization analysis of proteins and transcripts respectively (Heyland et al., 2011b). This work opened up several new avenues for my research, to combine life history theory with molecular methods as exemplified in my edited volume (Flatt and Heyland, 2011)further discussed below, as well as many of my other research and teaching efforts. It also led the way into the exploration of Aplysia neural peptide distribution in the CNS. For example my laboratory published a recent paper in the journal Endocrinology on the distribution and putative function of Glycoprotein hormone subunits (Heyland et al., 2012). Finally it helped my lab to develop a new research direction collaboratively with the Daphnia Genome Consortium at Indiana Bloomington, GLIER (The Great Lakes Institute for Environmental Research) at the University of Windsor (collaborator on NSERC CREATE program), Environment Canada and several industry partners. The focus of this collaborative effort is to gain a detailed molecular understanding of Daphniahomeostasis in fluctuating and stressful environments. This research has also led to the recent submission of a strategic grant where we propose to use waterfleas as a model organism for endocrine disrupting chemicals.
More Research in the Heyland Lab